FRASER Summary: all–v29
mumichae, vyepez, ischeller
2023-07-07
saveRDS(snakemake, snakemake@log$snakemake)
source(snakemake@params$setup, echo=FALSE)
suppressPackageStartupMessages({
library(cowplot)
library(RColorBrewer)
})dataset <- snakemake@wildcards$dataset
annotation <- snakemake@wildcards$annotation
padj_cutoff <- snakemake@config$aberrantSplicing$padjCutoff
deltaPsi_cutoff <- snakemake@config$aberrantSplicing$deltaPsiCutoff
fds <- loadFraserDataSet(file=snakemake@input$fdsin)
hasExternal <- length(levels(colData(fds)$isExternal) > 1)Number of samples: 100
Number of introns: 110839
Number of splice sites: 211180
# used for most plots
dataset_title <- paste0("Dataset: ", dataset, "--", annotation)Hyperparameter optimization
for(type in psiTypes){
g <- plotEncDimSearch(fds, type=type)
if (!is.null(g)) {
g <- g + theme_cowplot(font_size = 16) +
ggtitle(paste0("Q estimation, ", type)) + theme(legend.position = "none")
print(g)
}
}


Aberrantly spliced genes per sample
plotAberrantPerSample(fds, type=psiTypes,
padjCutoff = padj_cutoff, deltaPsiCutoff = deltaPsi_cutoff,
aggregate=TRUE, main=dataset_title) +
theme_cowplot(font_size = 16) +
theme(legend.position = "top")
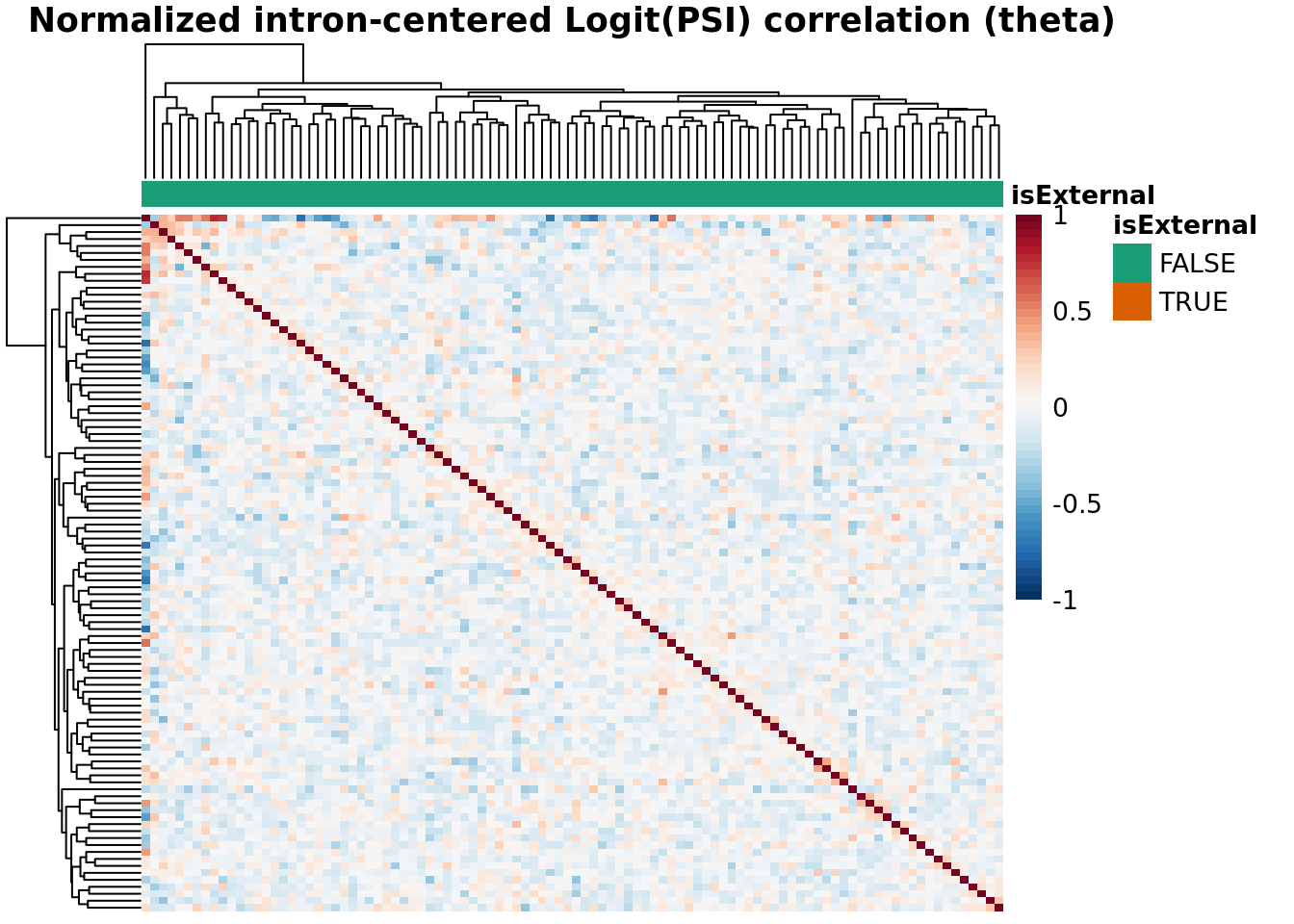
Batch Correlation: samples x samples
topN <- 30000
topJ <- 10000
anno_color_scheme <- brewer.pal(n = 3, name = 'Dark2')[1:2]
for(type in psiTypes){
for(normalized in c(F,T)){
hm <- plotCountCorHeatmap(
object=fds,
type = type,
logit = TRUE,
topN = topN,
topJ = topJ,
plotType = "sampleCorrelation",
normalized = normalized,
annotation_col = "isExternal",
annotation_row = NA,
sampleCluster = NA,
minDeltaPsi = minDeltaPsi,
plotMeanPsi = FALSE,
plotCov = FALSE,
annotation_legend = TRUE,
annotation_colors = list(isExternal = c("FALSE" = anno_color_scheme[1], "TRUE" = anno_color_scheme[2]))
)
hm
}
}
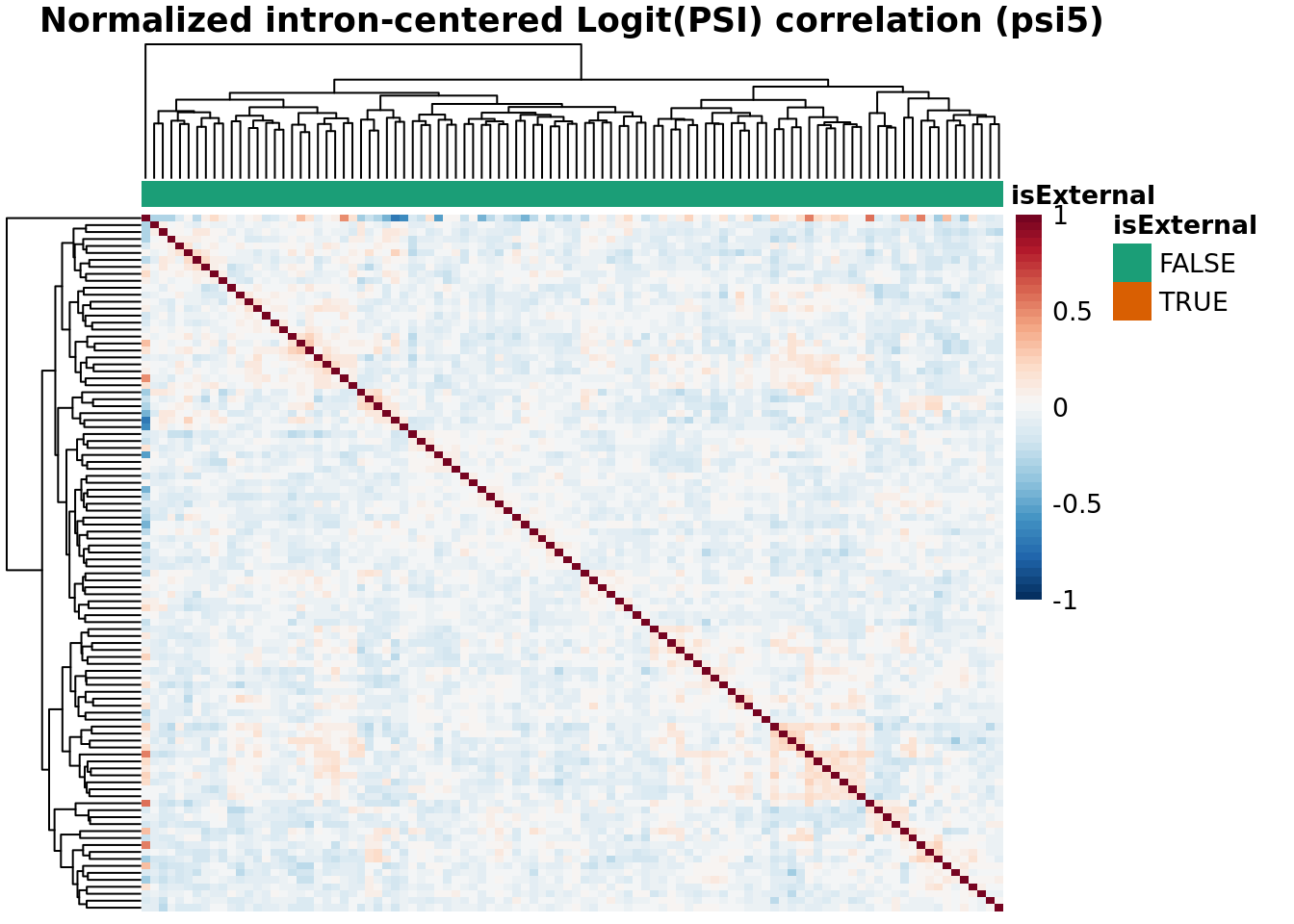
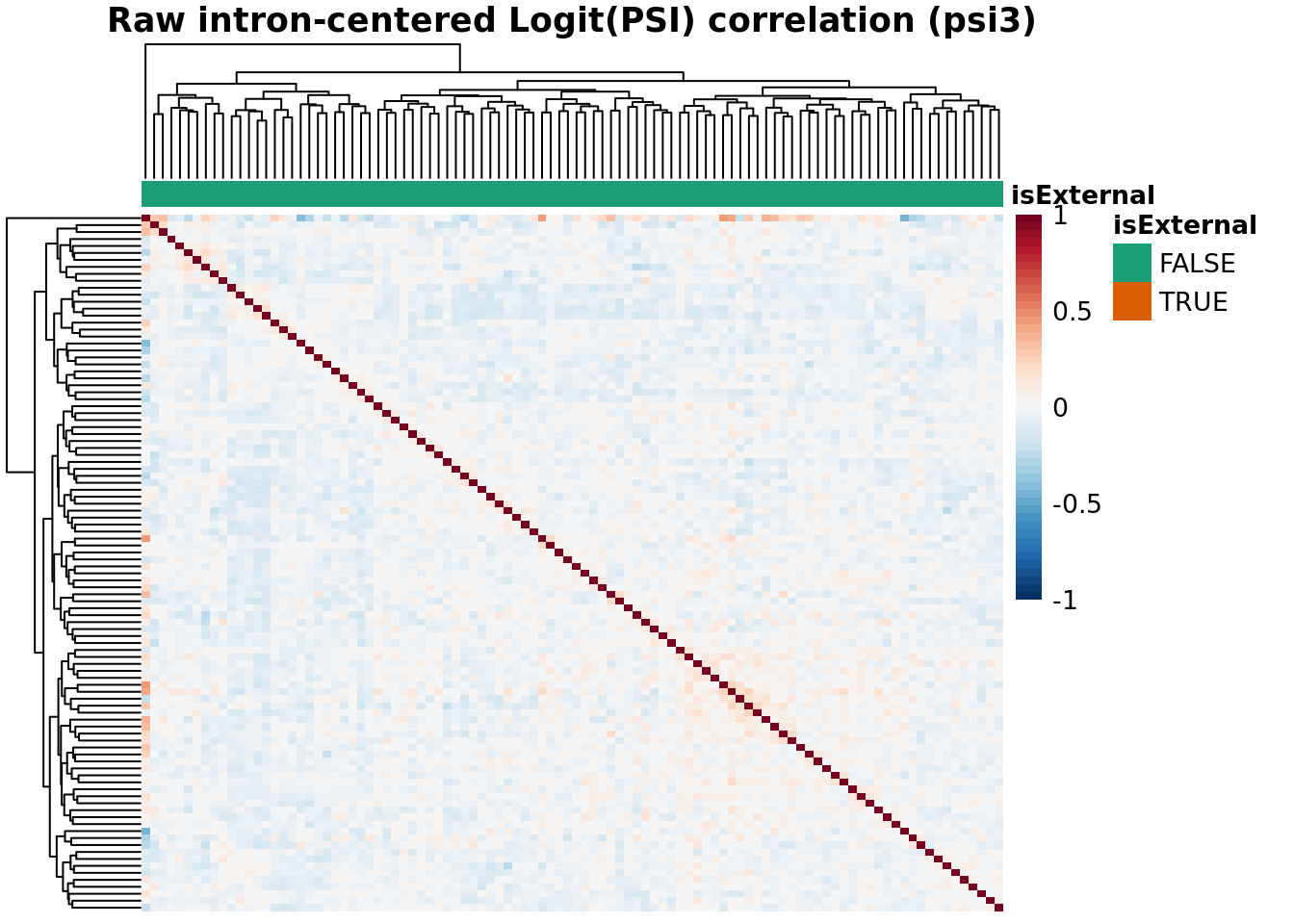
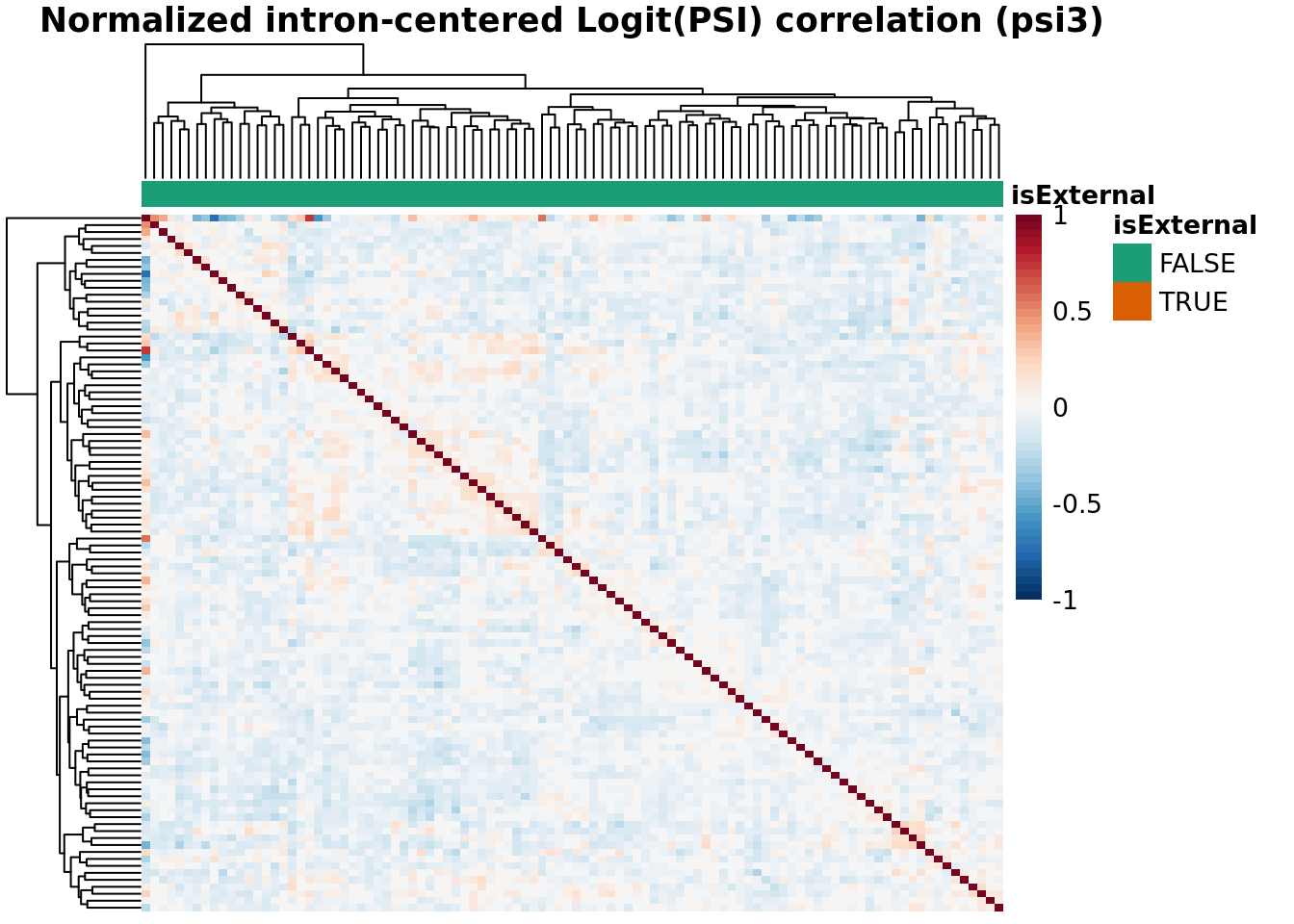
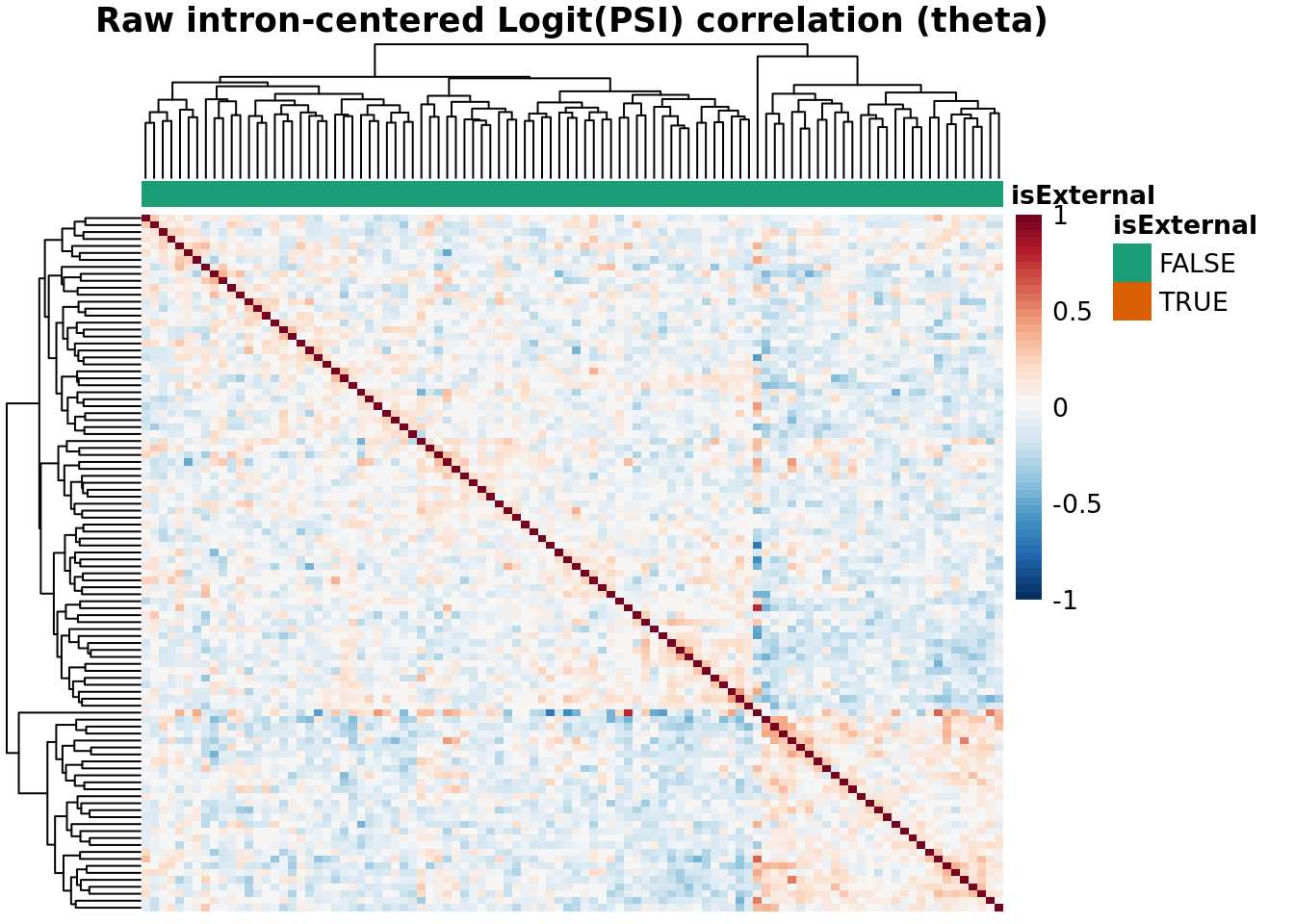
Results
res <- fread(snakemake@input$results)Total gene-level splicing outliers: 278
file <- snakemake@output$res_html
write_tsv(res, file = file)